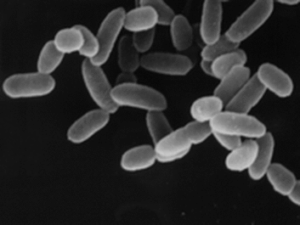
 Everyone has millions of microbes living in complex communities in their sinuses. All these hundreds of species of bacteria, fungi, and viruses are the sinus microbiome or sinus microbiota. In addition, some (many?) people also have tiny organisms called archaea living in their sinuses.
Everyone has millions of microbes living in complex communities in their sinuses. All these hundreds of species of bacteria, fungi, and viruses are the sinus microbiome or sinus microbiota. In addition, some (many?) people also have tiny organisms called archaea living in their sinuses.
What are archaea? Archaea are single-celled organisms that lack cell nuclei, and have a unique cell wall membrane. Very little is known about them, what their role is in the sinuses (that is, what are they doing there?), how do they interact with the host (the person), and whether their presence is beneficial or not.
There are only a few studies looking at archaea in humans, and while very little is known, the current view is that there are no known harmful archaea ("archaeal pathogens or parasites").
In a 2019 study, French researchers found archaea in the sinuses of 9 of their patients with chronic sinusitis - and therefore thought they were linked to disease. But unfortunately they didn't look to see if archaea are also found in the sinuses of healthy persons, thus there wasn't a comparison group. They found methanogenic archaea (the only microorganisms able to produce methane) in these nine patients, and they thought that the archaea were contributing to or causing the chronic sinusitis.
The Methanobrevibacter species they found were M. smithii, M. oralis, and M. massiliense, of which 2 have been found in dental plaque and periodontitis lesions, and one is a gut methanogen. [Note: This means it is found in the gut and is methane producing - but that doesn't mean it is harmful.]
Finally, a more recent and comprehensive study looked at archaea and bacteria in the sinuses of both healthy persons and those with chronic sinusitis. University of Auckland researchers found that only 6 out of 70 persons (both healthy and with sinusitis) had archaea in the sinuses, and they were very low in numbers and in diversity. In those with archaea, there was a lot of variation between people. They did not see any archaea associated with chronic sinusitis.
Archaea found were from Euryarchaeota, Thaumarchaeota, and Methanobrevibacteriaceae phyla.
One can only wonder what the archaea are doing in the sinuses in those with them. Especially, as the researchers point out that archaea are characterized by a unique cell wall membrane that "assists survival in extreme conditions such as hydrothermal vents, salt lakes, anoxic and highly acidic or alkaline environments". Also, that recent studies suggest that the human immune system recognizes and can be "activated" by archaea.

Finally, studies also mention that archaea are resistant to many antibiotics (because of lack of peptidoglycan in their cell wall). It is unknown how this influences their role (if any) in human health and disease.
As you can see, much is unknown right now. Even how many people have archaea in their sinuses, and what kinds of archaea. Stay tuned.
Article by B.W. Mackenzie et al in Frontiers in Cellular Infection and Microbiology: A Novel Description of the Human Sinus Archaeome During Health and Chronic Rhinosinusitis

 Something new to add to the list of what is in our skin microbiome - the community of microbes (bacteria, fungi, viruses) living on our skin. It turns out we also have archaea, which are
Something new to add to the list of what is in our skin microbiome - the community of microbes (bacteria, fungi, viruses) living on our skin. It turns out we also have archaea, which are 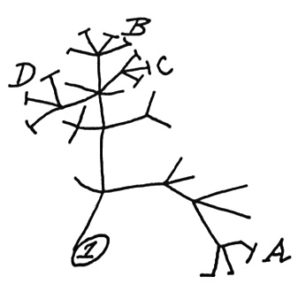 In 1837, Charles Darwin sketched a simple tree of life (shown left) to illustrate the idea that all living things share a common ancestor. Ever since then, scientists have been adding names to the
In 1837, Charles Darwin sketched a simple tree of life (shown left) to illustrate the idea that all living things share a common ancestor. Ever since then, scientists have been adding names to the 
 Clostridium difficile. Credit: CDC
Clostridium difficile. Credit: CDC