 The sinus microbiome consists of bacteria, viruses, and fungi all coexisting in the sinuses. Research has long focused on how the bacteria in sinus microbiomes in people with chronic sinusitis is different than in healthy persons without sinusitis (and yes, there are differences). Differences in the mycobiome (the fungi) of the sinuses appear to also play a role in chronic sinusitis.
The sinus microbiome consists of bacteria, viruses, and fungi all coexisting in the sinuses. Research has long focused on how the bacteria in sinus microbiomes in people with chronic sinusitis is different than in healthy persons without sinusitis (and yes, there are differences). Differences in the mycobiome (the fungi) of the sinuses appear to also play a role in chronic sinusitis.
A few recent studies found that there are differences in the fungi living in the sinuses in those without sinusitis and those with sinusitis (but results varied among the studies). Whether fungi play a role in the development of sinusitis has long been debated. Additionally, when samples are taken with a swab compared to sinus tissue samples during surgery - the results are different.
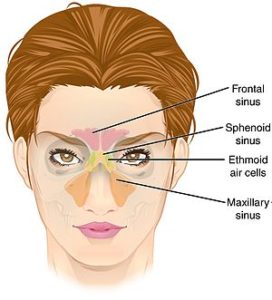
Credit: Nat. Library of Medicine
In a recent study, researchers found differences in both the fungi (mycobiome), as well as bacteria, in persons with chronic sinusitis compared to healthy persons (the controls). There were differences in bacterial diversity (diminished diversity in those with chronic sinusitis), as well as the composition of the microbes. The researchers thought that fungal alterations can play a part in sinusitis.
By the way, when microbes in the sinuses are out of whack it is called dysbiosis. Researchers point out that the same microbes can be commensal or pathogenic (if numbers increase to high numbers) or have no effect on sinusitis.
Excerpts from The Laryngoscope: Fungal and Bacterial Microbiome in Sinus Mucosa of Patients with and without Chronic Rhinosinusitis
Abstract Objectives - Dysbiosis of the sinonasal microbiome has been implicated in the pathogenesis of chronic rhinosinusitis (CRS). However, the mycobiome remains largely understudied, and microbial alterations associated with specific CRS subtypes have yet to be delineated. The objective of this study is to investigate the fungal and bacterial microbiome of sinus mucosa in CRS patients with and without nasal polyposis (CRSwNP and CRSsNP) versus healthy controls.
Methods - Sinus mucosa was obtained from 92 patients (31 CRSsNP, 31 CRSwNP, and 30 controls) undergoing endoscopic sinus/skull base surgery.... Fungal and bacterial microbiome analysis was performed utilizing internal transcribed spacer amplicon and 16S rRNA sequencing.
Results - Beta diversity of the sinonasal mycobiome differed significantly between CRS and controls and between CRSwNP and controls, but not between CRSwNP and CRSsNP nor between CRSsNP and controls. With respect to the bacterial microbiome, significantly lower alpha diversity was observed between CRS and controls , CRSwNP versus controls, and CRSsNP versus controls.
Conclusions - Differences in mycobiota diversity in CRS patients in comparison with controls suggest that alterations in the mycobiome may contribute to disease pathogenesis. Our findings also confirmed that diminished diversity among bacterial communities is associated with CRS and that significant differences are present in microbial composition between CRSwNP and CRSsNP.
Recent studies have demonstrated that relative microbial abundance is an impactful measure of disease; and reduced diversity with overgrowth of certain species, even if initially commensal, may be associated with CRS....Although fungal dysbiosis has been reported in CRS, other studies have found the prevalence and composition of fungi to be similar among patients with/without CRS [chronic rhinosinusitis]
Overall, similar to the bacterial microbiome, our findings suggest that reduced fungal diversity may be important in the mechanism of dysbiosis-driven CRS, but at this point, the degree of dysbiosis is not noticeably different between CRSwNP and CRSsNP.
In the current study, a higher abundance of Alternaria, a dermatiaceous fungus associated with allergic rhinosinusitis, and lower abundance of Cutaneotrichosporon were detected among CRS versus controls.
With respect to CRS subgroups, the mycobiome composition was not significantly different between controls CRSsNP or CRSwNP, but there were notable differences in the abundance of several microbes between CRSwNP and controls. Saccharomycetales and Cutaneotrichosporon were lower among CRSwNP, and Alternaria species were higher among CRSwNP.
In terms of microbial composition and abundance in CRS, prior reports have been highly variable. Corynebacterium, Pseudomonas, Staphylococcus, etc. have been considered commensal in some studies, whereas others found the same microbes as well as Streptococcus, Haemophilus, Moraxella, etc. to be pathogenic.9-19 ... In the current study, we also identified a wide array of organisms with significantly different abundances in CRS samples compared with controls, including Corynebacteriaceae and Pseudomonadaceae. ... However, as reflected by the varied results in the literature, the roles of microbes in CRS likely change depending on the complex interplay between individual and environmental factors, as different studies have found the same organism to be commensal, pathogenic, or have no role in CRS.
