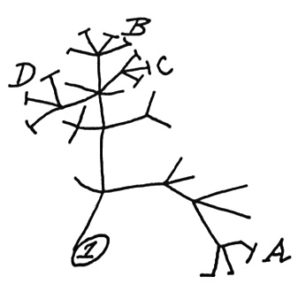
 In 1837, Charles Darwin sketched a simple tree of life (shown left) to illustrate the idea that all living things share a common ancestor. Ever since then, scientists have been adding names to the tree of life, including a massive effort of 2.3 million named species of animals, plants, fungi and microbes in 2015. A tree of life is a visual hypothesis of how scientists think species are related to one another, so it has been evolving over the years as more information is learned and species discovered.
In 1837, Charles Darwin sketched a simple tree of life (shown left) to illustrate the idea that all living things share a common ancestor. Ever since then, scientists have been adding names to the tree of life, including a massive effort of 2.3 million named species of animals, plants, fungi and microbes in 2015. A tree of life is a visual hypothesis of how scientists think species are related to one another, so it has been evolving over the years as more information is learned and species discovered.
Now a group of 17 researchers have sketched out a radically different tree of life. It has two main trunks—one full of bacteria and another comprised of archaea, a group of single-celled microbes that run on very different biochemistry. The eukaryotes—the domain that includes all animals, fungi, and plants—are crowded on a thin branch coming off the archaeal trunk. About half of these bacterial branches belong to a supergroup, which was discovered very recently and is currently known as the candidate phyla radiation. Within these branches are numerous species that we’re almost completely ignorant about, and they’ve never been isolated or grown in a lab (with one exception called TM7). In fact, this supergroup of bacteria and “other lineages that lack isolated representatives clearly comprise the majority of life’s current diversity,” write researchers Hug and Banfield. Wow. From Science Daily:
Wealth of unsuspected new microbes expands tree of life
Scientists have dramatically expanded the tree of life, which depicts the variety and evolution of life on Earth, to account for over a thousand new microscopic life forms discovered over the past 15 years. The expanded view finally gives bacteria and Archaea their due, showing that about two-thirds of all diversity on Earth is bacterial -- half bacteria that cannot be isolated and grown in the lab -- while nearly one-third is Archaeal.
Much of this microbial diversity remained hidden until the genome revolution allowed researchers like Banfield to search directly for their genomes in the environment, rather than trying to culture them in a lab dish. Many of the microbes cannot be isolated and cultured because they cannot live on their own: they must beg, borrow or steal stuff from other animals or microbes, either as parasites, symbiotic organisms or scavengers.
The new tree, to be published online April 11 in the new journal Nature Microbiology, reinforces once again that the life we see around us -- plants, animals, humans and other so-called eukaryotes -- represent a tiny percentage of the world's biodiversity.
"Bacteria and Archaea from major lineages completely lacking isolated representatives comprise the majority of life's diversity," said Banfield.....According to first author Laura Hug,... the more than 1,000 newly reported organisms appearing on the revised tree are from a range of environments, including a hot spring in Yellowstone National Park, a salt flat in Chile's Atacama desert, terrestrial and wetland sediments, a sparkling water geyser, meadow soil and the inside of a dolphin's mouth. All of these newly recognized organisms are known only from their genomes.
One striking aspect of the new tree of life is that a group of bacteria described as the "candidate phyla radiation" forms a very major branch. Only recognized recently, and seemingly comprised only of bacteria with symbiotic lifestyles, the candidate phyla radiation now appears to contain around half of all bacterial evolutionary diversity.
Charles Darwin first sketched a tree of life in 1837 as he sought ways of showing how plants, animals and bacteria are related to one another. The idea took root in the 19th century, with the tips of the twigs representing life on Earth today, while the branches connecting them to the trunk implied evolutionary relationships among these creatures.....Archaea were first added in 1977 after work showing that they are distinctly different from bacteria, though they are single-celled like bacteria. A tree published in 1990 by microbiologist Carl Woese was "a transformative visualization of the tree," Banfield said. With its three domains, it remains the most recognizable today.
With the increasing ease of DNA sequencing in the 2000s, Banfield and others began sequencing whole communities of organisms at once and picking out the individual groups based on their genes alone. This metagenomic sequencing revealed whole new groups of bacteria and Archaea, many of them from extreme environments, such as the toxic puddles in abandoned mines, the dirt under toxic waste sites and the human gut. Some of these had been detected before, but nothing was known about them because they wouldn't survive when isolated in a lab dish.
For the new paper, Banfield and Hug teamed up with more than a dozen other researchers who have sequenced new microbial species, gathering 1,011 previously unpublished genomes to add to already known genome sequences of organisms representing the major families of life on Earth.....The analysis, representing the total diversity among all sequenced genomes, produced a tree with branches dominated by bacteria, especially by uncultivated bacteria. A second view of the tree grouped organisms by their evolutionary distance from one another rather than current taxonomic definitions, making clear that about one-third of all biodiversity comes from bacteria, one-third from uncultivable bacteria and a bit less than one-third from Archaea and eukaryotes. (Original article and diagrams.)


 Clostridium difficile. Credit: CDC
Clostridium difficile. Credit: CDC