 This research supports the growing evidence for the importance of microbes in the health of young children. Even the type of milk a child drinks is important. From Science Daily:
This research supports the growing evidence for the importance of microbes in the health of young children. Even the type of milk a child drinks is important. From Science Daily:
Pediatric allergology: Fresh milk keeps infections at bay
A study by researchers of Ludwig-Maximilians-Universitaet (LMU) in Munich shows that infants fed on fresh rather than UHT (ultra-pasteurized) cow's milk are less prone to infection.
A pan-European study, led by Professor Erika von Mutius, Professor of Pediatric Allergology at LMU and Head of the Asthma and Allergy Department at Dr. von Hauner's Children's Hospital, reports that fresh cow's milk protects young children from respiratory infections, febrile illness and inflammation of the middle ear. As untreated cow's milk may itself contain pathogenic microorganisms and could pose a health risk, the researchers argue for the use of processing methods that preserve the protective agents present in raw milk.
The findings are the latest to emerge from the long-term PASTURE study, which is exploring the role of dietary and environmental factors in the development of allergic illness. The study initially recruited 1000 pregnant women who were asked to document their children's diet and state of health at weekly intervals during the first year of life.
"Among children who were fed on fresh, unprocessed cow's milk the incidence of head colds and other respiratory infections, febrile and middle-ear inflammation was found to be significantly lower than in the group whose milk ration consisted of the commercially processed ultra-pasteurized product," says Dr. Georg Loss. Ingestion of farm milk reduced the risk of developing these conditions by up to 30%, and the effect was diminished if the milk was heated at home before consumption. Conventionally pasteurized milk retained the ability to reduce the risk of febrile illness, while exposure to the higher temperatures used in UHT processing eliminated the effect altogether.
"The effects of diverse milk treatments are presumably attributable to differentially heat-resistant components present in fresh milk. Compounds that are sensitive to heating seem to play a particularly important role in protection against respiratory-tract and ear infections," says Loss.
At the end of the first year of life, blood samples were obtained from the children enrolled in the study, and tested for biochemical indicators of immunological function. Infants fed on unprocessed milk were found to have lower levels of the C-reactive protein, which is a measure of inflammation status. "Other studies have shown that higher levels of inflammation are related to the subsequent emergence of chronic conditions such as asthma and obesity. Consumption of unprocessed milk may therefore reduce the risk of developing asthma," Loss explains.
Industrial processing of milk involves short-term heating of the raw product. Conventionally pasteurized milk has been exposed to temperatures of 72-75°C for 15 seconds, while ultra-pasteurized milk undergoes heating at around 135°C for a few seconds. The latter is also homogenized to disperse the milk fats, which prevents the formation of cream.
In addition to fats and carbohydrates, cow's milk contains proteins that can modulate the function of the immune system. "In many respects, the composition of cow's milk is similar to that of human milk," says Loss. It has long been known that breast-feeding protects infants from infection, although how milk actually affects the early immune function remains unclear. It is possible that some of the factors involved interact directly with viruses or that they promote the development of a healthy immune system by altering the composition of the gut microflora.
That living in the country has positive effects on the immune system has been demonstrated in several previous studies. Together these investigations show, as Erika von Mutius notes, that "children who grow up on traditional dairy farms are least likely to develop allergies.

 Very exciting new way to use probiotics! Huge potential. From Science Daily:
Very exciting new way to use probiotics! Huge potential. From Science Daily: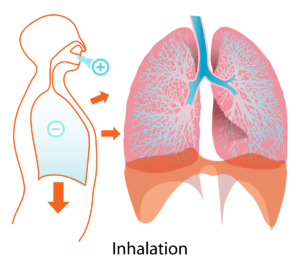 Human lungs. Credit: Wikipedia
Human lungs. Credit: Wikipedia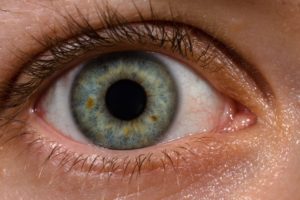 Finally, a discussion of the eye microbiome or microbiota (microbial community). Originally eyes were thought not to have much microbial life. But now with modern technology (such as genetic sequencing) it is known that many microbial species live on the eye. And yes, the eye or ocular microbiome can become imbalanced.
Finally, a discussion of the eye microbiome or microbiota (microbial community). Originally eyes were thought not to have much microbial life. But now with modern technology (such as genetic sequencing) it is known that many microbial species live on the eye. And yes, the eye or ocular microbiome can become imbalanced.