 This article discusses the fungi living on our skin. Recent research (using state of the art genetic analysis) has found that healthy people have lots of diversity in fungi living on their skin. Certain areas seem to have the greatest populations of fungi: in between toes (average of 40 species), the heel (average of 80 species), toenails (average of 80 species), and the genitals. Currently it is thought that there are "intricate interactions between fungi and immune cells on the skin surface", and that often this mutualistic relationship is beneficial, but at other times dysbiosis (when the microbial community is unbalanced or out of whack) can lead to diseases. If the populations get too unbalanced (e.g., antibiotics can kill off bacteria, and then an increase in fungi populations take their place) then ordinarily non-harmful fungi can become pathogenic. Note that: Mutualistic relationship is a relationship between two different species of organisms in which both benefit from the association. From E-Cronicon:
This article discusses the fungi living on our skin. Recent research (using state of the art genetic analysis) has found that healthy people have lots of diversity in fungi living on their skin. Certain areas seem to have the greatest populations of fungi: in between toes (average of 40 species), the heel (average of 80 species), toenails (average of 80 species), and the genitals. Currently it is thought that there are "intricate interactions between fungi and immune cells on the skin surface", and that often this mutualistic relationship is beneficial, but at other times dysbiosis (when the microbial community is unbalanced or out of whack) can lead to diseases. If the populations get too unbalanced (e.g., antibiotics can kill off bacteria, and then an increase in fungi populations take their place) then ordinarily non-harmful fungi can become pathogenic. Note that: Mutualistic relationship is a relationship between two different species of organisms in which both benefit from the association. From E-Cronicon:
From Head to Toe: Mapping Fungi across Human Skin
The human microbiota refers to the complex aggregate of fungi, bacteria and archaea, found on the surface of the skin, within saliva and oral mucosa, the conjunctiva, the gastrointestinal. When microbial genomes are accounted for, the term microbiome is deployed. In recent years the first in-depth analysis, using sophisticated DNA sequencing, of the human microbiome has taken place through the U.S. National Institutes of Health led Human Microbiome Project.
Many of the findings have extended, or even turned upside down, what was previously known about the relationship between humans and microorganisms. One of the most interesting areas related to fungi, especially in advancing our understanding about fungal types, locations and numbers and how this affects health and disease....some parts of the body have a greater prevalence of bacteria (such as the arms) whereas fungi are found in closer association with feet.
A variety of bacteria and fungi are found on the typical 2 square meters that represent the surface of the skin, and within the deeper layers, of a typical adult. These can be considered as ‘residential’ (that is ordinarily found) or ‘transient’ (carried for a period of time by the host.) The resident microorganism types vary in relation to skin type on the human body; between men and women; and to the geographical region in which people live.
The first observation is that many locations across the skin contain considerable populations of fungi. Prime locations, as reported by Findley and colleagues, were inside the ear canal and behind the ear, within the eyebrows, at the back of the head; with feet: on the heel, toenails, between the toes; and with the rest of the body notable locations were the forearm, back, groin, nostrils, chest, palm, and the elbow.
The second observation is that several different species are found, and these vary according to different niches. Focusing on one ecological niche, a study by Oyeka found that the region between toes, taken from a sample of 100 people, discovered 14 genera of fungi. In terms of the individual species recovered, a relatively high number were observed (an average of 40 species.)....the greatest varieties of fungi are to be found on the heel (approximately 80 different species.) The second most populous area is with the toes, where toe nails recover around 80 different species.....With the genitals, where early investigations had suggested that Candida albicans was the most commonly isolated yeasts. However, an investigation of 83 patients by Bentubo., et al. showed more variety, with high recoveries of Candida parapsilosis, Rhodotorulamucilaginos, Rhodotorulaglutinis, Candida tropicalis and Trichosporoninkin.
The importance of the investigative work into the human skin fungi helps medical researchers understand more fully the connections between the composition of skin-fungi and certain pathologies. Here the intricate interactions between fungi and immune cells on the skin surface is of importance; often this mutualistic relationship is beneficial, at other times dysbiosis can lead to the manifestation of diseases especially when there is a breakdown of the mutualistic relationship.
Changes to fungal diversity can be associated with several health conditions, including atopic dermatitis, psoriasis, acne vulgaris and chronic wounds. Diversity can alter through the over-use of antibiotics, where a decline in bacterial numbers can lead to a rise in fungal populations occupying the same space.
Moreover, research has indicted that patients who have a primary immunodeficiency are host to more populous fungal communities than healthy people. Here it is suggested that the weaknesses in the immune system allow higher numbers of fungi to survive, and, in turn these weaknesses can lead some ordinarily non-harmful species to become pathogenic. Such opportunistic fungi include species of Aspergillus and Candida.
 The following article is interesting because it describes how microbes are high up in the sky riding air currents and winds to circle the earth, and eventually drop down somewhere. This is one way diseases can be spread from one part of the world to another. And the study looking at how antibiotic resistant bacteria are spread in the air from cattle feedlots has implications for how antibiotic resistance is spread. From Smithsonian:
The following article is interesting because it describes how microbes are high up in the sky riding air currents and winds to circle the earth, and eventually drop down somewhere. This is one way diseases can be spread from one part of the world to another. And the study looking at how antibiotic resistant bacteria are spread in the air from cattle feedlots has implications for how antibiotic resistance is spread. From Smithsonian:
 Nice update from a large crowd sourced study I posted about
Nice update from a large crowd sourced study I posted about  There has been tremendous concern in recent years over pathogenic bacteria (such as Salmonella and Escherichia coli) found on raw fruits and vegetables. But what about nonpathogenic bacteria? Aren't some of the benefits of eating raw fruits and vegetables the microbes found on them? What actually is on them?
There has been tremendous concern in recent years over pathogenic bacteria (such as Salmonella and Escherichia coli) found on raw fruits and vegetables. But what about nonpathogenic bacteria? Aren't some of the benefits of eating raw fruits and vegetables the microbes found on them? What actually is on them? Amazing! We each release a "personal microbial cloud" with its own "microbial cloud signature" every day. The unique combination of millions of bacteria (from our microbiome or community of microbes - including bacteria, viruses, fungi - that live within and on us) can identify us. Not only do we each give off a unique combination, but we each give off different amounts of microbes - some more, some less. Some very common bacteria: Streptococcus, Propionobacterium, Corynebacterium, and Lactobacillus (among women).The microbes are given off with every movement, every exhalation, every scratching of the head, every burp and fart, etc. - and they go in the air around the person and settle around the person (they researchers even collected bacteria from dishes set on the ground around the person). From Science Daily:
Amazing! We each release a "personal microbial cloud" with its own "microbial cloud signature" every day. The unique combination of millions of bacteria (from our microbiome or community of microbes - including bacteria, viruses, fungi - that live within and on us) can identify us. Not only do we each give off a unique combination, but we each give off different amounts of microbes - some more, some less. Some very common bacteria: Streptococcus, Propionobacterium, Corynebacterium, and Lactobacillus (among women).The microbes are given off with every movement, every exhalation, every scratching of the head, every burp and fart, etc. - and they go in the air around the person and settle around the person (they researchers even collected bacteria from dishes set on the ground around the person). From Science Daily: This article discusses the fungi living on our skin. Recent research (using state of the art genetic analysis) has found that healthy people have lots of diversity in fungi living on their skin. Certain areas seem to have the greatest populations of fungi:
This article discusses the fungi living on our skin. Recent research (using state of the art genetic analysis) has found that healthy people have lots of diversity in fungi living on their skin. Certain areas seem to have the greatest populations of fungi:  Every time you inhale, you suck in thousands of microbes. And depending on where you live, the microbes will vary. From Wired:
Every time you inhale, you suck in thousands of microbes. And depending on where you live, the microbes will vary. From Wired: This nice general summary of what scientists know about the microbial community within us was just published by a division of the NIH (National Institutes of Health). Very simple and basic. From the National Institute of General Medical Sciences (NIGMS):
This nice general summary of what scientists know about the microbial community within us was just published by a division of the NIH (National Institutes of Health). Very simple and basic. From the National Institute of General Medical Sciences (NIGMS):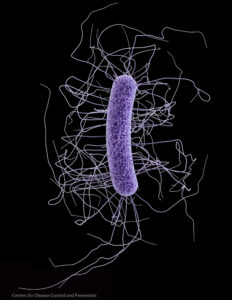 Clostridium difficile. Credit: CDC
Clostridium difficile. Credit: CDC