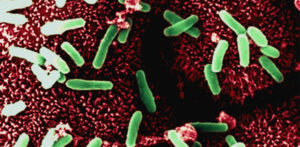
 The American Academy of Pediatrics released a new report that the overuse of antibiotics in animals poses a real health risk to children. Giving routine antibiotics to animals leads to antibiotic resistant bacteria - which means that antibiotics may not work when given to people. Most of the antibiotics sold in the U.S. each year - 80 percent- are used in animals that people than eat. The great majority of antibiotics given to animals are the same ones given to humans.The main way to ensure that the meat that you are purchasing is antibiotic-free is to buy meat labeled organic. And to buy organic dairy products (milk, butter, cheese, cream). Note that the reason routine use of antibiotics in animals has not been stopped so far in the USA is due to agriculture industry lobbying. From Medical Xpress:
The American Academy of Pediatrics released a new report that the overuse of antibiotics in animals poses a real health risk to children. Giving routine antibiotics to animals leads to antibiotic resistant bacteria - which means that antibiotics may not work when given to people. Most of the antibiotics sold in the U.S. each year - 80 percent- are used in animals that people than eat. The great majority of antibiotics given to animals are the same ones given to humans.The main way to ensure that the meat that you are purchasing is antibiotic-free is to buy meat labeled organic. And to buy organic dairy products (milk, butter, cheese, cream). Note that the reason routine use of antibiotics in animals has not been stopped so far in the USA is due to agriculture industry lobbying. From Medical Xpress:
Pediatricians' group urges cuts in antibiotic use in livestock
Overuse of antibiotics in farm animals poses a real health risk to children, the American Academy of Pediatrics warns in a new report.This common practice is already contributing to bacterial resistance to medicines and affecting doctors' ability to treat life-threatening infections in kids, according to the paper published online Nov. 16 in the journal Pediatrics.
"The connection between production uses of antibiotics in the agricultural sector to antibiotic resistance is alarming," said Victoria Richards, an associate professor of medical sciences at the Quinnipiac University School of Medicine in Hamden, Conn. She believes the danger is "not only for infants and children but other vulnerable populations, such as the pregnant and the older individuals."
As the academy explained in its warning, antibiotics are often added to the feed of healthy livestock to boost growth, increase feed efficiency or prevent disease. However, the practice can also make antibiotics ineffective when they are needed to treat infections in people. Some examples of emerging antibiotic germs include methicillin-resistant staphylococcus aureus (MRSA), C.difficile, and highly resistant strains of the tuberculosis bacterium. Each year, more than 2 million Americans develop antibiotic-resistant infections and more than 23,000 die from these infections, the academy said. And in 2013, the highest incidence of such infections was among children younger than 5, federal government statistics show.
"Children can be exposed to multiple-drug resistant bacteria, which are extremely difficult to treat if they cause an infection, through contact with animals given antibiotics and through consuming the meat of those animals," report author Dr. Jerome Paulson, immediate past chair of the academy's executive committee of the Council on Environmental Health, said in an academy news release."Like humans, farm animals should receive appropriate antibiotics for bacterial infections," he said. "However, the indiscriminate use of antibiotics without a prescription or the input of a veterinarian puts the health of children at risk."
Spaeth noted that the U.S. Centers for Disease Control and Prevention, as well as the World Health Organization, have both called for a curbing of antibiotic use in animals. But the authors of the new report expressed concern over resistance from the agriculture and farming industry to such measures.

 New research found that one course of
New research found that one course of  Nice update from a large crowd sourced study I posted about
Nice update from a large crowd sourced study I posted about  Could this be? Fungal infection being the cause of Alzheimer's disease? Noteworthy from a recent study conducted in Spain: all the Alzheimer's disease (AD) patients had evidence of fungal infections in their brains, central nervous systems, and vascular systems, but none were found in the control subjects (those without Alzheimer's disease). Many of the symptoms of AD (such as inflammation of the central nervous system and activation of the immune system) match those with long-lasting fungal infections. A "microbial cause" has long been suggested as a cause of AD, and interestingly other studies have also found fungal infections in AD patients. The research so far has found
Could this be? Fungal infection being the cause of Alzheimer's disease? Noteworthy from a recent study conducted in Spain: all the Alzheimer's disease (AD) patients had evidence of fungal infections in their brains, central nervous systems, and vascular systems, but none were found in the control subjects (those without Alzheimer's disease). Many of the symptoms of AD (such as inflammation of the central nervous system and activation of the immune system) match those with long-lasting fungal infections. A "microbial cause" has long been suggested as a cause of AD, and interestingly other studies have also found fungal infections in AD patients. The research so far has found 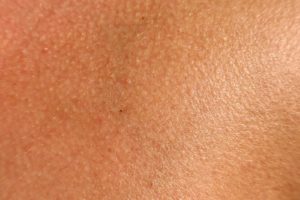 The microbes living on healthy human skin include bacteria, fungi, and viruses...but 90% of the viruses found on healthy skin in this study are unknown to researchers - thus "
The microbes living on healthy human skin include bacteria, fungi, and viruses...but 90% of the viruses found on healthy skin in this study are unknown to researchers - thus " According to a new report, it looks like most people under the age of 50 (throughout the world) have herpes simplex virus infections - whether type 1 or type 2. (Picture is of a herpes simplex virus type 1, at www.virology.net). From Medical Xpress:
According to a new report, it looks like most people under the age of 50 (throughout the world) have herpes simplex virus infections - whether type 1 or type 2. (Picture is of a herpes simplex virus type 1, at www.virology.net). From Medical Xpress: There has been tremendous concern in recent years over pathogenic bacteria (such as Salmonella and Escherichia coli) found on raw fruits and vegetables. But what about nonpathogenic bacteria? Aren't some of the benefits of eating raw fruits and vegetables the microbes found on them? What actually is on them?
There has been tremendous concern in recent years over pathogenic bacteria (such as Salmonella and Escherichia coli) found on raw fruits and vegetables. But what about nonpathogenic bacteria? Aren't some of the benefits of eating raw fruits and vegetables the microbes found on them? What actually is on them? An interesting Canadian study that followed young children for 3 years found that young infants may be more likely to develop allergic
An interesting Canadian study that followed young children for 3 years found that young infants may be more likely to develop allergic  This study found something surprising in many samples of human breast tissue -
This study found something surprising in many samples of human breast tissue -  Amazing! We each release a "personal microbial cloud" with its own "microbial cloud signature" every day. The unique combination of millions of bacteria (from our microbiome or community of microbes - including bacteria, viruses, fungi - that live within and on us) can identify us. Not only do we each give off a unique combination, but we each give off different amounts of microbes - some more, some less. Some very common bacteria: Streptococcus, Propionobacterium, Corynebacterium, and Lactobacillus (among women).The microbes are given off with every movement, every exhalation, every scratching of the head, every burp and fart, etc. - and they go in the air around the person and settle around the person (they researchers even collected bacteria from dishes set on the ground around the person). From Science Daily:
Amazing! We each release a "personal microbial cloud" with its own "microbial cloud signature" every day. The unique combination of millions of bacteria (from our microbiome or community of microbes - including bacteria, viruses, fungi - that live within and on us) can identify us. Not only do we each give off a unique combination, but we each give off different amounts of microbes - some more, some less. Some very common bacteria: Streptococcus, Propionobacterium, Corynebacterium, and Lactobacillus (among women).The microbes are given off with every movement, every exhalation, every scratching of the head, every burp and fart, etc. - and they go in the air around the person and settle around the person (they researchers even collected bacteria from dishes set on the ground around the person). From Science Daily: